SARS-CoV-2 Assembled Genome
Coronavirus disease 2019 (COVID-19) has been rapidly spreading across the world since its first identification. This unprecedented pandemic has led to the tragic loss of human life and has presented a colossal challenge to public health research. The disease is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), whose genome is of great importance to the elucidation of viral dynamics and the search for a proper treatment.
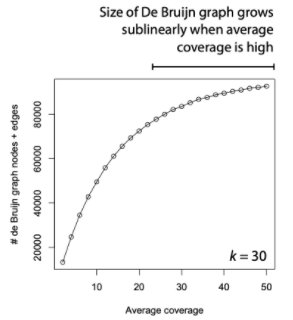
Assembling short reads from sequencing techniques into the correct order is a fundamental task of bioinformatics research. A number of methods have been developed over the past few decades, among which Euler path based techniques have proved efficient and reliable. In this project, we construct De Bruijn graphs from SAS-Cov-2 whole genome sequencing reads and implement an algorithm to establish an Euler path in each graph to explore the factors, such as the length of K-mers, that might affect genome assembly. In addition, we explore methods to visualize the graph and validate our results with a reference genome. Furthermore, we report the results of our tentative approaches to comparing the impact of different lengths of K-mers and the results of the De Bruijn graph construction and visualization.
If you would like to read more about our results and findings, please visit our GitHub Repository.

Contribution
- Implement the De Bruijn graph and the Eulerian path/circuit
- With the Eulerian path, we report the assembled contigs by first picking a starting node. If a node has an extra outgoing edge, we will use it. Otherwise, we can pick any random node, and then traverse through the entire graph by using depth-first traversal and backtracking to discover unvisited edges. Once it is done, we combine all the reported nodes back together.
- Assist the group to write and present the discovery/finding reports of the COVID-19 assembled genome
- Filter and clean up FASTQ raw data
- Correct sequencing errors for the shotgun sequencing
- Break sequences/k-mers to use the De Bruijn graph
- Use the Eulerian path to report the assembled genome
- Make unit tests for the Graph and the Eulerian path
Lessons Learned
- Algorithms can help us understand and solve complex problems to save lives
- Learning from others’ skills and backgrounds is important when working on a team
- Communication is key since so each member can fully understand the goals and strategies in a project
Possible Future Work
- Implement a balancing algorithm to take every node to have the same number of outgoing edges with the incoming edges so the Eulerian circuit exists